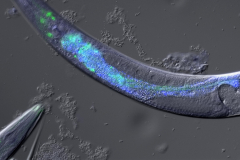
Projects
Stress response in C. elegans:
C. elegans is a well-established model organism for studying stress response mechanisms. It can respond to a variety of stressors, including temperature fluctuations (heat and cold), oxidative stress, hypoxia, radiation, and infection, by activating a complex network of signaling pathways and transcriptional regulators.
One of the key signaling pathways involved in the C. elegans stress response is the insulin/IGF-1 signaling pathway, which is conserved across many species. Under stress conditions, this pathway is inhibited, leading to the activation of downstream effectors such as the FOXO transcription factor DAF-16. DAF-16, in turn, upregulates the expression of stress response genes, including those involved in heat shock protein production, oxidative stress defense, and DNA damage repair.
Another critical pathway is the mechanistic target of rapamycin (mTOR) pathway. The two mTOR complexes, mTORC1 and mTORC2, have distinct roles in stress regulation. While mTORC1 promotes growth and proliferation while suppressing stress responses, mTORC2 enhances cellular resilience and supports stress adaptation.
Additional pathways contributing to stress responses in C. elegans include the unfolded protein response (UPR), which is triggered by the accumulation of misfolded proteins in the endoplasmic reticulum, and the p38 MAPK pathway, which is activated by various stressors such as heat shock and oxidative stress.
C. elegans is a well-established model organism for studying stress response mechanisms. The worm can respond to various stressors, including temperature (heat, cold), oxidative stress, hypoxia, radiation, and infection, by activating a complex network of signaling pathways and transcriptional regulators.
One of the key signaling pathways involved in stress response in C. elegans is the insulin/IGF-1 signaling pathway, which is conserved across many species. In response to stress, this pathway is inhibited, activating downstream effectors such as the FOXO transcription factor DAF-16. DAF-16 then activates the expression of stress response genes, including those involved in heat shock protein production, oxidative stress response, and DNA damage repair.
Another important pathway involved in stress response in C. elegans is the mTOR (mechanistic target of rapamycin) pathway. the two mTOR complexes, mTORC1 and mTORC2, have distinct functions and regulation. While mTORC1 promotes growth and proliferation and suppresses the stress response, mTORC2 has been shown to enhance the stress response and promote cellular resilience to stress.
Other pathways involved in stress response in C. elegans include the unfolded protein response (UPR), which is activated in response to the accumulation of misfolded proteins in the endoplasmic reticulum, and the p38 MAPK pathway, which is activated in response to various stressors, including heat shock and oxidative stress.
Mitochondrial stress responses: Using genome-wide screenings, we have identified 55 candidate genes that contribute to the mitochondrial stress response upon application of reactive oxygen species that impair mitochondrial electron transport chain activities. We have proposed mechanisms how the mitochondria respond to oxidative stress through activating mitochondrial chaperone synthesis, and have identified a kinase that links endoplasmic and mitochondrial stress responses. Our work on mitochondrial stress also translates to a better understanding of genes involved in neurodegenerative disorders, most notably Parkinson’s Disease.
Cross talk between insulin and TOR signaling in tumorigenesis and stress response: Using both animal models (C. elegans) and cell culture, we identified several new functional connectivities of insulin/IGF and TOR signaling with relevance to human cancer. In particular, we identified novel regulators and interactors of mTORC using large-scale mass spectrometry approaches, and a previously unknown crosstalk between mTORC and TGFß signaling.
Insulin signalling in tumorigenesis: We study stem cell tumorigenesis (hyperplasia, cell migration and tissue invasion) using the nematode C. elegans and human cell cultures as models. In detail, we have established worm models of germline stem cell tumors encompassing tumor-suppressive, but also oncogenic properties of the FOXO transcription factor DAF-16, a major downstream effector of insulin/IGF (IIs) signaling in worms and humans. During the past funding period of BIOSS, we have described novel oncogenic, non-cell-autonomous activities of FOXO and have identified novel inhibitors and enhancers of tumorigenesis, including genes that function in pathways that crosstalk with IIs. We have also found that dysfunction of the tumor suppressor gene PTEN/DAF-18 may result in stem cell tumors, depending on an earlier nutritional status of the organism.
Signaling function of Parkinson’s Disease genes: We have shown functional relationships of several genes, whose mutations segregate with hereditary Parkinson’s Disease. Most notably, we showed antagonistic activities of PINK1 and LRRK2 and their C. elegans orthologues, and have identified signaling mechanisms to understand their functional relationships. Our findings that intracellular membrane transport is diversely affected by PD genes have allowed us screening for modulators of PD gene dysfunction, the results of which we currently investigating.
Methods
C. elegans disease/signaling models: The work-horse of our studies is the model organism Caenorhabditis elegans, which allows us to set up assays for the functional characterization of novel components of signaling pathways, evaluation of gene function, the generation of synthetic models encompassing a living animal system supplemented with human genes of interest that integrate seemingless.
CRISPR/Cas9 genome editing: We have developed several protocols that allow us successfully to generate genomic deletions (gene knock-outs), but also single base-pair alterations of genomic regions (genome editing, e.g. to change single amino acids in a protein) in C. elegans. This method has been used to systematically introduce loss-of-function and gain-of-function (e.g. constitutively active proteins, or to generate mutations at positions implicated with hereditary diseases) mutations in the genome of C. elegans. Thus, we generated for example point mutations resulting in amino acid exchanges of proteins homologous to mutants involved in human disease. We are currently expanding these protocols to allow integration of reporter genes at dedicated sites of the genome, to generate in vivo tagged proteins, or to genomically integrate entire human genes under the control of a C. elegans promoter.
FRAP: A Powerful Method to Evaluate Membrane Fluidity in C. elegans: FRAP (Fluorescence Recovery After Photobleaching) is probably the most direct method to investigate the dynamics of molecules in living cells. We use FRAP to analyze membrane fluidity in different mutants and different conditions which influences the fluidity of cellular membranes in certain mutants.
Synthetic organisms for pharmacological drug screenings: We can generate synthetic organisms that allow drug screenings against human proteins/mutants in the functional environment of an entire organism. We have used a variety of methods, e.g. to integrate human Parkinson’s Disease variants into the worm, and have used this synthetic organism to screen for drugs ameliorating mutant phenotypes. Our lab has experience in screenings for up to 6,000 compounds in medium-scale settings, but also ample experience in upscaling such screens to higher throughput.
FACS-like sorting of entire organisms: We run a FACS-like C. elegans sorter that allows us high-throughput sorting of living specimens according to several criteria: length, density, structural abnormalities, expression of marker proteins, fluorescence, lipid content etc. Dual sortings and resortings, as well as subsequent distribution of candidates to microtiter plates are also possible.
Proximity proteomics and labelling: Methods for the in vivo labeling of protein nano-environments. We are employing proximity proteomics approaches that were first introduced into the BIOSS portfolio by Michael Reth’s group, with which we have established several collaborations. For tandem-tag of proteins, our lab has developed the Snavi-Tag system that we have made available to the public through ADDGENE.
Genome-wide screenings for modulators of gene function: Our main model organism C. elegans is well-known for genome-wide screenings, e.g. to identify novel pathway components, modifiers of mutant dysfunction, alterations of behavior and more. We own several libraries for RNA interference of basically every C. elegans gene and have used these libraries successfully to identify novel gene functions and interactions. Moreover, we generated sublibraries of e.g. all kinases and all transcription factors encoded in the genome. Using our bioinformatics platform, rapid generation of novel sub-libraries, e.g. phosphatase libraries, or GPCR genes, are easily doable.
Behavioral analyses: C. elegans is a model very well-known to couple neurobiological functions to even complex behavioral outputs, including motility, mating behavior, learning paradigms, sensation of olfactory, chemical, mechanical, osmotic environmental signals. We have a tool-box of more than 150 phenotypic readouts that we can employ to even address complex functions of nervous system or whole organisms.
Stress biology and endomembrane transport dynamics: We have invested in the last years in many tools to monitor mitochondrial dynamics and functions, and to investigate endomembrane transport properties, lysosomes and ER functions, protein aggregation and ROS stress, intracellular sorting dynamics, necrosis and apoptosis, phagocytosis and autophagy. Our toolbox includes models in both C. elegans tissues and human cell cultures.
Interactor screenings: Our group has implemented tools to study protein interactions using the split-ubiquitin system that allows identification of binary or tertiary interactions in the living cell. This method has been stream-lined to particularly allow monitoring interactions of transcription factors and of membrane-bound proteins, interactions of the latter are notoriously difficult to analyze. We have established or acquired proteome libraries for screenings of human brain, human fetal brain, kidney, different stages and conditions of C. elegans, as well as of other model organisms, including plants. Automation of these procedures have been initiated in collaboration with ZBSA and the University of Marburg. Moreover, we have established protein interaction screenings using mass spectrometry tools. These have been used extensively in the past year to identify novel interactors of human stress signaling and metabolic pathways, and of C. elegans proteins.